 Parallelized ultra-high throughput microfluidicDroplet-based microfluidic technologies are powerful tools for applications requiring high-throughput, for example, in biochemistry or material sciences. Several systems have been proposed for the high-throughput production of monodisperse emulsions by parallelizing multiple droplet makers. However, these systems have two main limitations: (1) they allow the use of only a single disperse phase; (2) they are based on multiple layer microfabrication techniques. We present here a pipette-and-play solution offering the possibility of manipulating simultaneously 10 different disperse phases on a single layer device. This system allows high-throughput emulsion production using aqueous flow rates of up to 26 ml/h (>110 000 drops/s) leading to emulsions with user-defined complex chemical composition. We demonstrate the multiplex capabilities of our system by measuring the kinetics of β-galactosidase in droplets using nine different concentrations of a fluorogenic substrate.
|
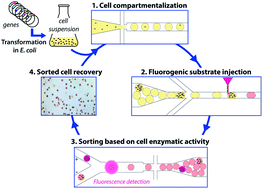 CotA laccase: high-throughput manipulation and analysis of recombinant enzyme libraries expressed in E. coli using droplet-based microfluidics
We present a high-throughput droplet-based microfluidic analysis/screening platform for directed evolution of CotA laccase: droplet-based microfluidic modules were combined to develop an efficient system that allows cell detection and sorting based on the enzymatic activity. This platform was run on two different operating modes: the "analysis" mode allowing the analysis of the enzymatic activity in droplets at very high rates (>1000 Hz) and the "screening" mode allowing sorting of active droplets at 400 Hz. The screening mode was validated for the directed evolution of the cytoplasmic CotA laccase from B. subtilis, a potential interesting thermophilic cathodic catalyst for biofuel cells. Single E. coli cells expressing either the active CotA laccase (E. coli CotA) or an inactive frameshifted variant (E. coli ΔCotA) were compartmentalized in aqueous droplets containing expression medium. After cell growth and protein expression within the droplets, a fluorogenic substrate was "picoinjected" in each droplet. Fluorescence-activated droplet sorting was then used to sort the droplets containing the desired activity and the corresponding cells were then recultivated and identified using colorimetric assays. We demonstrated that E. coli CotA cells were enriched 191-fold from a 1:9 initial ratio of E. coli CotA to E. coli ΔCotA cells (or 437-fold from a 1:99 initial ratio) using a sorting rate of 400 droplets per s. This system allows screening of 106 cells in only 4 h, compared to 11 days for screening using microtitre plate-based systems. Besides this low error rate sorting mode, the system can also be used at higher throughputs in "enrichment" screening mode to make an initial purification of a library before further steps of selection. Analysis mode, without sorting, was used to rapidly quantify the activity of a CotA library constructed using error-prone PCR. This mode allows analysis of 106 cells in only 1.5 h.
|
 Ultra-high throughput detection of single cell β-galactosidase activity in droplets using micro-optical lens array
We demonstrate the use of a hybrid microfluidic-micro-optical system for the screening of enzymatic activity at the single cell level. Escherichia coli β-galactosidase activity is revealed by a fluorogenic assay in 100 pl droplets. Individual droplets containing cells are screened by measuring their fluorescence signal using a high-speed camera. The measurement is parallelized over 100 channels equipped with microlenses and analyzed by image processing. A reinjection rate of 1 ml of emulsion per minute was reached corresponding to more than 105 droplets per second, an analytical throughput larger than those obtained using flow cytometry.
|
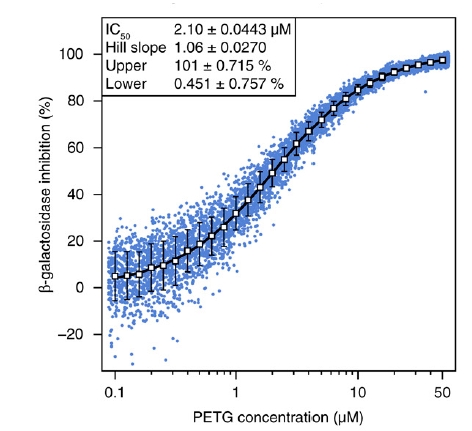 Droplet-based microfluidics for drug screening applicationsIn the quest for new therapeutic agents, the pharmaceutical industry is reaching the limits of existing screening technologies, such as the automatized microtitre plates. The current state of the art enable automatized assays at rates up to 1 test per second on volumes of about 1 microlitre for 1 concentration of the compound to be tested. The activity of a molecule being a function of its concentration, in principle, a perfect screening system would require the test of different concentrations for a given molecule but it is clear that multiplying the conditions of screening is just adding up in the total screening time and expense in reagents.
There is therefore a need for the fast and automatized manipulation of small amount of chemicals for drug screening applications. In this respect droplet-based microfluidics offers very promising alternatives.
We have shown here that such microfluidic platforms can be designed for the screening of potential drugs from microtitre plates in small droplets of a few picolitre and enable the automated measurements of dose-response curve. We use here a basic hydrodynamic principle (the Taylor-Aris dispersion) to generate the concentration gradient of the chemical before encapsulation in the droplets. The activity of the chemical is then assayed on therapeutic targets and revealed by a fluorogenic assay. By recording the fluorescence of a dye encoding the drug concentration and the fluorescence readout of the assay, automatized dose-response curves are measured. The screening plateform has been integrated with an autosampler for the screening of compound libraries. The high resolution of the data reveals complex dose-response relationships unambiguously. We used this system to screen a chemical library of 704 compounds against protein tyrosine phosphatase 1B, a diabetes, obesity, and cancer target and identified a number of novel inhibitors, the most potent being sodium cefsulodine, which has an IC50 of about 30 microM.
This project was done in a collaboration with the company Raindance Technologies (USA) and the pharmaceutical company Sanofi.
High-resolution dose-response screening using droplet-based microfluidics
O.J. Miller, A. El Harrak, T. Mangeat, J.-C. Baret, L. Frenz, B. El Debs, E. Mayot, M.L. Samuels, E.K. Rooney, P. Dieu, M. Galvan, D.R. Link and A.D. Griffiths
Proc. Natl. Acad. Sci. U S A 10 (2012) 378-383
| |
|
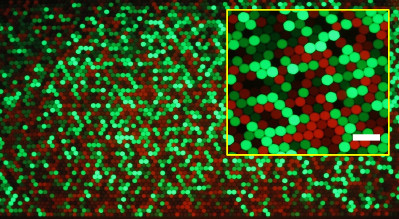
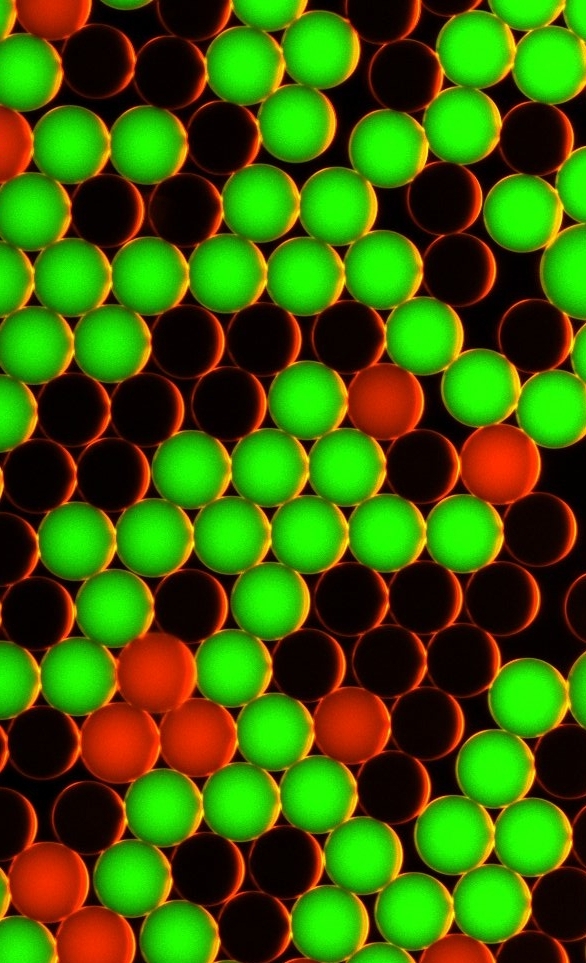 Counting fluorescent droplets for the detection of cancer biomarkersComplex networks of genetic alterations have been highlighted for most cancers. These somatic mutations are present in tumor cells but not in normal cells and can serve as highly specific biomarkers. They represent molecular signatures of cancer cells and thus constitute great tools for early cancer detection, for prognostic assessment, and also for the detection of possible recurrence. The ability to detect such biomarkers would enable to define which patient could benefit from a treatment, and therefore - considering the large number of possible treatment - to treat each patient with the most efficient treatment combination for his own cancer.
Target-oriented treatments have met increasing developments in the last decade and new biomarkers of resistance or sensitivity to such treatments have been described. However, the techniques used nowadays in clinical oncology enable the detection of biomarkers when they represent a fraction of at least 10 % of the normal DNA, while DNA from tumors can be present at less than 0,1% in blood samples. Plain use of the biomarkers in clinical oncology requires then highly sensitive and specific methods to detect them accurately in clinical samples. We considered the case of somatic KRAS mutations, found at high rates in Leukemia, colon cancer, pancreatic cancer and lung cancer; KRAS mutation is a predictive factor of resistance for some therapies and 90% of the activating mutations occur at 7 DNA positions in 2 codons (12 and 13). We have developed an original procedure to quantify the exact fraction of mutant DNA in a biological sample down to 1 KRAS mutant sequence in 200 000 non mutated sequence, where oncologists could not currently detect less than 1 in 10.
This method combines the latest developments of molecular diagnostics with microfluidic manipulation of droplets in microfluidic chips. The droplets have a size of a few microns and are produced and manipulated in microchannels of several tens of microns (smaller than the diameter of an hair). The DNA is partitioned in the microdroplets, with no more than one gene copy per droplet, together with two fluorescent probes: a green one which recognizes the mutated gene, and a red one, which recognizes the normal gene. A droplet containing initially a normal gene will fluoresce in red, a droplet initially containing a mutant gene will fluoresce in green (a droplet containing none will not fluoresce). The simple counting of red and green droplet (which can be automatically done over millions of droplets in microfluidics) gives the ratio of mutants to normal genes. This work has been performed in a collaboration between the Université de Strasbourg, the Université Paris-Descartes, the Max-Planck Institute for Dynamics and Self-organization and the american company Raindance Technologies and could constitute a new paradigm for cancer diagnosis. These tools, protocols and procedures could, for example, be used for appropriate patient management by identifying which patient are more likely to benefit from specific targeted treatment or detecting in a non-invasive way Cancer recurrence in various bodily fluids.
Quantitative and sensitive detection of rare mutations using droplet-based microfluidics
D. Pekin, Y. Skhiri, J.-C. Baret, D. Le Corre, L. Mazutis, C. Ben Salem, F. Millot, A. El Harrak, J.B. Hutchison, J.W. Larson, D.R. Link, P. Laurent-Puig, A.D. Griffiths and V. Taly
Lab Chip 11, 2156-2166 (2011)
| |
Droplet-based Microfluidics: Sensitive Detection of Rare Mutations, J.C. Baret and V. Taly
|
CollaborationsDr. Valerie Taly, ISIS, University of Strasbourg / CNRS (France)
Prof. Andrew Griffiths, ISIS, University of Strasbourg / CNRS (France)
Raindance Technologies (USA) |
Related Publications
High-resolution dose-response screening using droplet-based microfluidics
O.J. Miller, A. El Harrak, T. Mangeat, J.-C. Baret, L. Frenz, B. El Debs, E. Mayot, M.L. Samuels, E.K. Rooney, P. Dieu, M. Galvan, D.R. Link and A.D. Griffiths
Proc. Natl. Acad. Sci. U S A 10 (2012) 378-383
| |
Quantitative and sensitive detection of rare mutations using droplet-based microfluidics
D. Pekin, Y. Skhiri, J.-C. Baret, D. Le Corre, L. Mazutis, C. Ben Salem, F. Millot, A. El Harrak, J.B. Hutchison, J.W. Larson, D.R. Link, P. Laurent-Puig, A.D. Griffiths and V. Taly
Lab Chip 11, 2156-2166 (2011)
| |
Fluorescence-activated droplet sorting (FADS): efficient microfluidic cell sorting based on enzymatic activity
J.-C. Baret, O. J. Miller, V. Taly, M. Ryckelynck, A. El-Harrak, L. Frenz, C. Rick, M. L. Samuels, J. B. Hutchison, J. J. Agresti, D. R. Link, D. A. Weitz, and A. D. Griffiths
Lab Chip 9, 1850 (2009)
| |
Droplet-based microfluidic systems for high-throughput single DNA molecule isothermal amplification and analysis
L. Mazutis, A. F. Araghi, O. J. Miller, J.-C. Baret, L. Frenz, A. Janoshazi, V. Taly, B. J. Miller, J. B. Hutchison, D. Link, A. D. Griffiths, and M. Ryckelynck
Anal. Chem. 81, 4813 (2009)
| |
|







